本文来自爱可可的前沿推介
推荐理由:研究展示了用生成式人工智能在零样本模式下生成新抗体变体的能力,生成具有天然序列特征和高度多样性的结合体,有可能颠覆传统的抗体药物发现方法,节省大量时间和成本的同时,提供更可控的设计选项。
Unlocking de novo antibody design with generative artificial intelligence
A Shanehsazzadeh, S Bachas, G Kasun, J M. Sutton
[Absci Corporation]
用生成人式工智能解锁新抗体设计
要点:
-
用生成式深度学习模型以零样本方式设计了针对三种不同目标的抗体; -
筛查了40万个抗体变体,发现三个结合比治疗抗体 trastuzumab 更紧密; -
生成的序列具有高度的多样性,与已知抗体序列相似性低,具有可变的结构形态; -
设计方法高度可控,可创建优化可开发性和免疫原性特征的蛋白质,减少下游可开发性风险。
摘要:
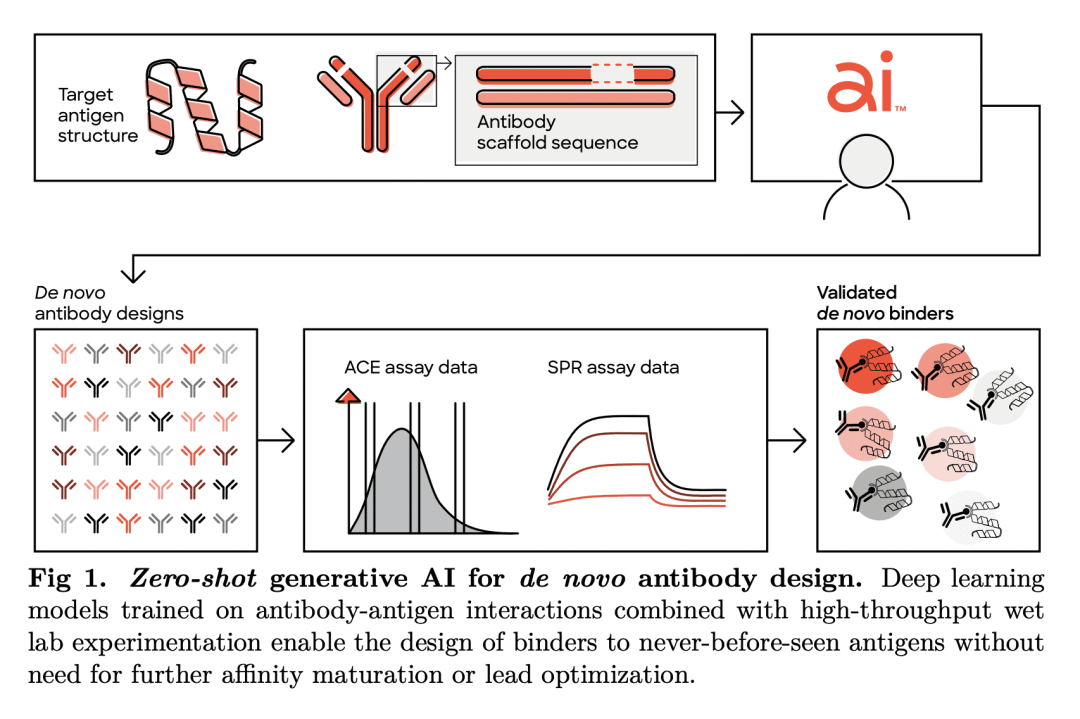
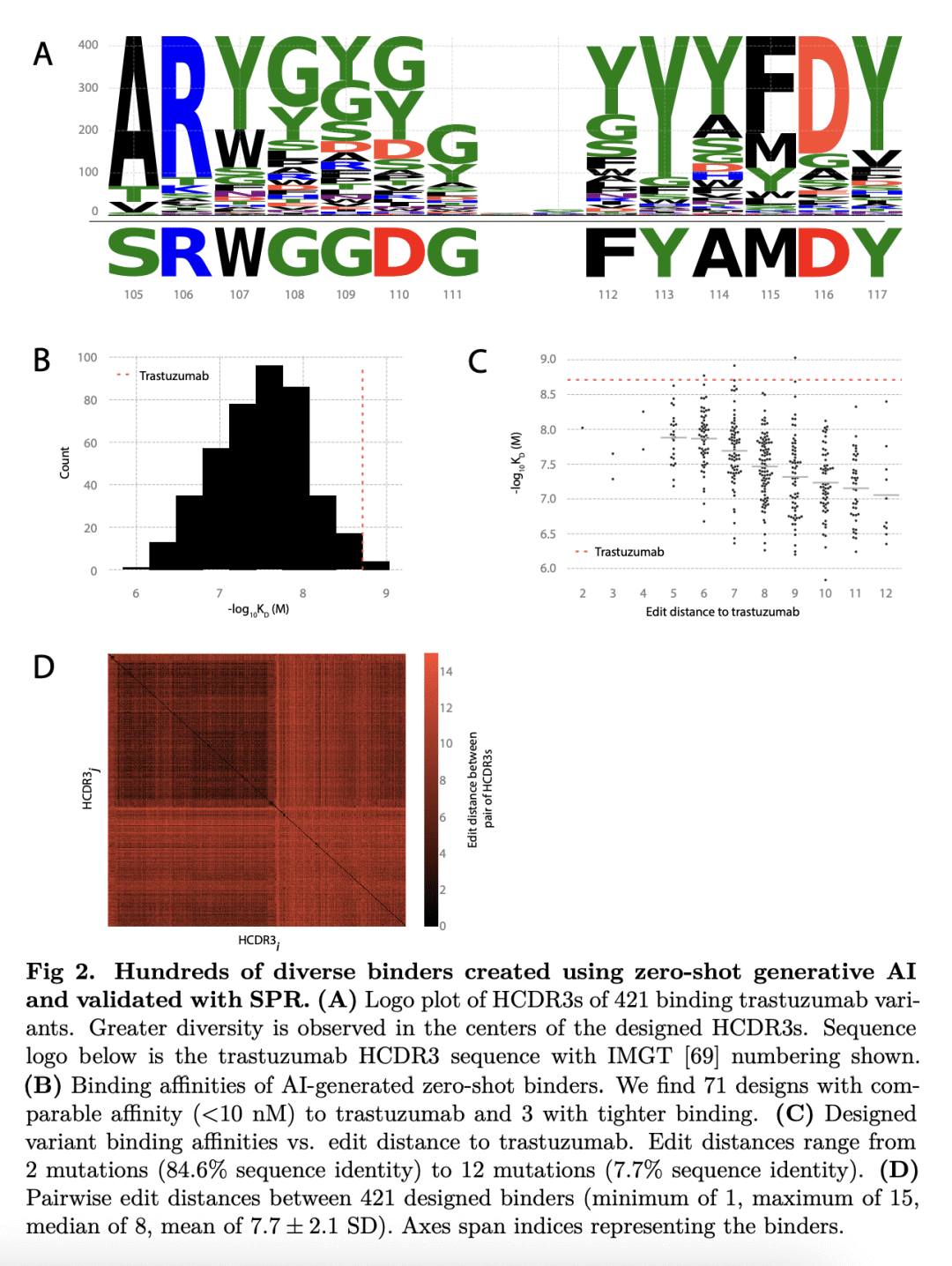
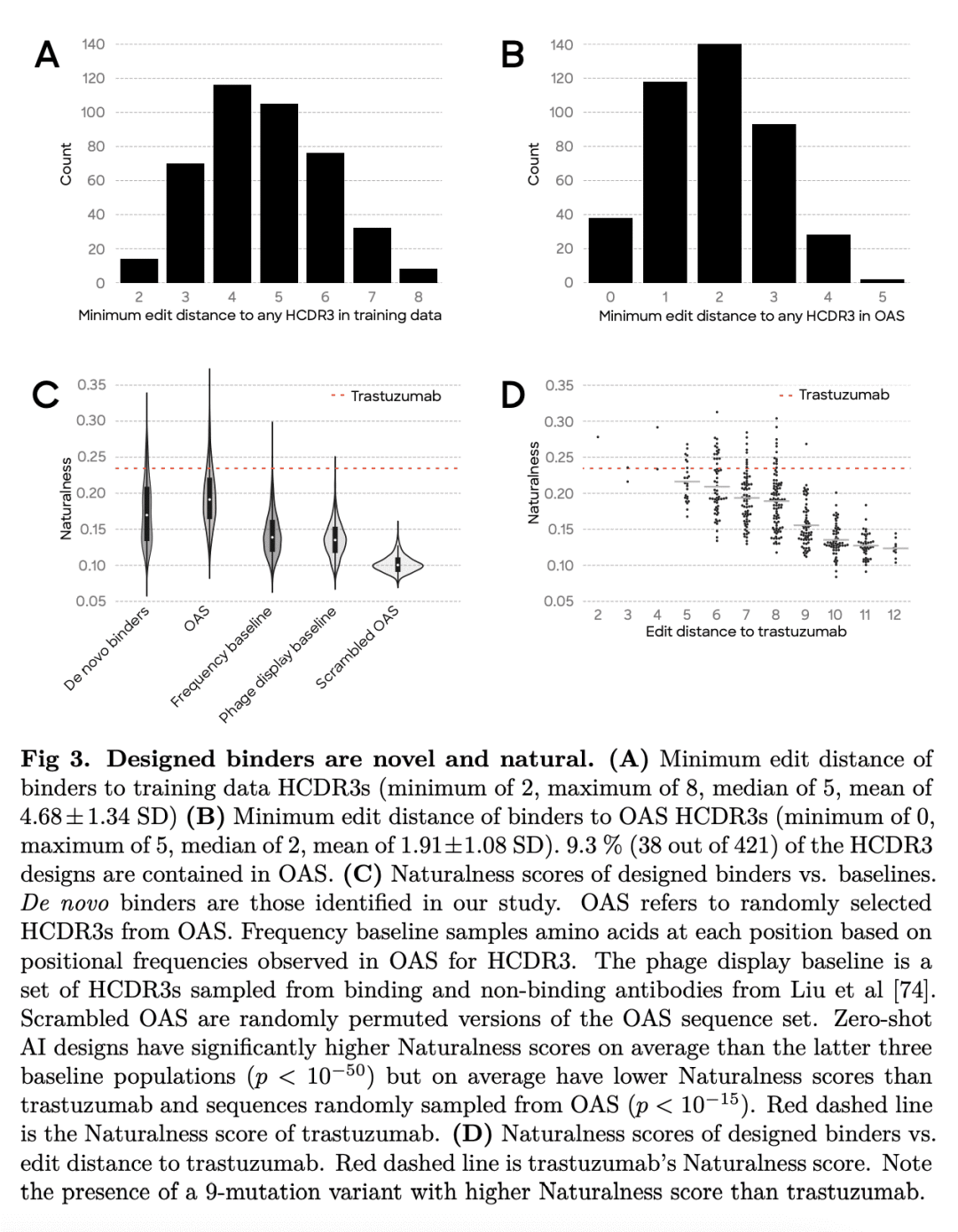
生成人工智能(AI)有可能大大提高抗体设计的速度、质量和可控性。传统的新抗体发现,需要对大型免疫或合成库进行时间和资源密集型筛查。这些方法对输出序列的控制也很少,这可能导致具有次优绑定和不良可开发性属性的候选者胜出。有人引入了具有硅基证据前景的生成抗体设计模型,然而,该方法没有通过实验验证以证明新的抗体设计。本文采用生成式深度学习模型,以零样本方式针对三个不同目标设计抗体,所有设计都是一轮模型生成的结果,没有后续优化。特别的,筛选了40多万种旨在与人类表皮生长因子受体2(HER2)结合的抗体变体。用表面等离子体共振(SPR)进一步表示了421个粘合剂,发现三种比治疗性抗体曲妥珠单抗结合得更紧密的粘合剂。粘合剂高度多样化,对已知抗体的序列恒等性较低,并采用可变结构构象。这些结果为使用生成式人工智能和高通量实验为新治疗目标加速药物创造开辟了道路。
论文地址:https://www.biorxiv.org/content/10.1101/2023.01.08.523187v1
Generative artificial intelligence (AI) has the potential to greatly increase the speed, quality and controllability of antibody design. Traditional de novo antibody discovery requires time and resource intensive screening of large immune or synthetic libraries. These methods also offer little control over the output sequences, which can result in lead candidates with sub-optimal binding and poor developability attributes. Several groups have introduced models for generative antibody design with promising in silico evidence [1–10], however, no such method has demonstrated de novo antibody design with experimental validation. Here we use generative deep learning models to de novo design antibodies against three distinct targets, in a zero-shot fashion, where all designs are the result of a single round of model generations with no follow-up optimization. In particular, we screen over 400,000 antibody variants designed for binding to human epidermal growth factor receptor 2 (HER2) using our high-throughput wet lab capabilities. From these screens, we further characterize 421 binders using surface plasmon resonance (SPR), finding three that bind tighter than the therapeutic antibody trastuzumab. The binders are highly diverse, have low sequence identity to known antibodies, and adopt variable structural conformations. Additionally, these binders score highly on our previously introduced Naturalness metric [11], indicating they are likely to possess desirable developability profiles and low immunogenicity. We open source1 the HER2 binders and report the measured binding affinities. These results unlock a path to accelerated drug creation for novel therapeutic targets using generative AI combined with high-throughput experimentation.

内容中包含的图片若涉及版权问题,请及时与我们联系删除



评论
沙发等你来抢